Probabilistically-weighted RNA-Seq Alignments
Colin Dewey, PhD – Cancer Genetics Program
Jamie Thomson, PhD – Cell Signaling Program
- Key to RNA-seq is accurate mapping of reads to reference genome.
- Earlier read-mapping algorithms threw out reads that mapped to multiple sites.
- RSEM assigns probability distribution of sites for each read and refines by expectation-maximization
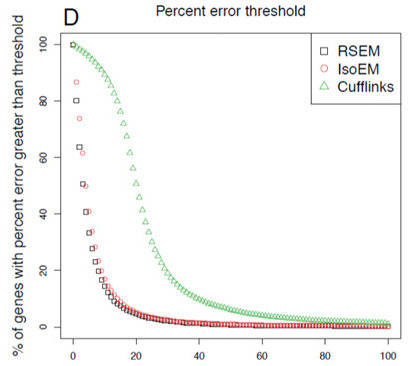
- Goal is to have lowest-curve few genes with high error in expression measurement
- RSEM outperforms competitors.
Mammography – Machine Learning
Elizabeth Burnside, MD, PhD – Cancer Prevention & Control Program
C. David Page, PhD – Cancer Genetics Program
- Goal is to accurately identify malignant abnormalities on mammogram.
- Data entered in National Mammography Database standard format.
- Ground truth malignant/benign from breast cancer registry, based on biopsy.
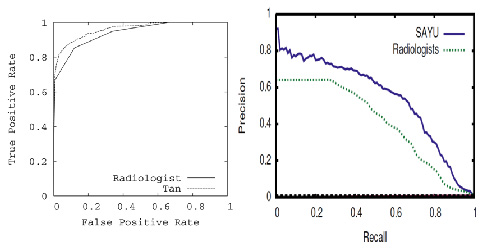
- Bayes net learning algorithm yields model with significantly better ROC area on held-out data than radiologists.
- New learning algorithm SAYU performs even better.
- Precision-recall curves are another way to plot ROC data for rare events and show room for further improvement.
Analysis of RNASeq Data
CISR supported Dr. Bresnick (GEM) and colleagues in illustrating exosome complex’s role during erythropoiesis. This work discovered that exosome complex is a vital determinant of a developmental signaling transition that dictates red blood cell proliferation versus differentiation. CISR staff scientist Peng Liu processedExosc3-/-ES cells RNA-seq data, performed differential gene expression analysis, and quantified the change of Kit expression levels, revealing Kit expression is 5.9-fold lower in Exosc3-/- ES cells than in normal ones.
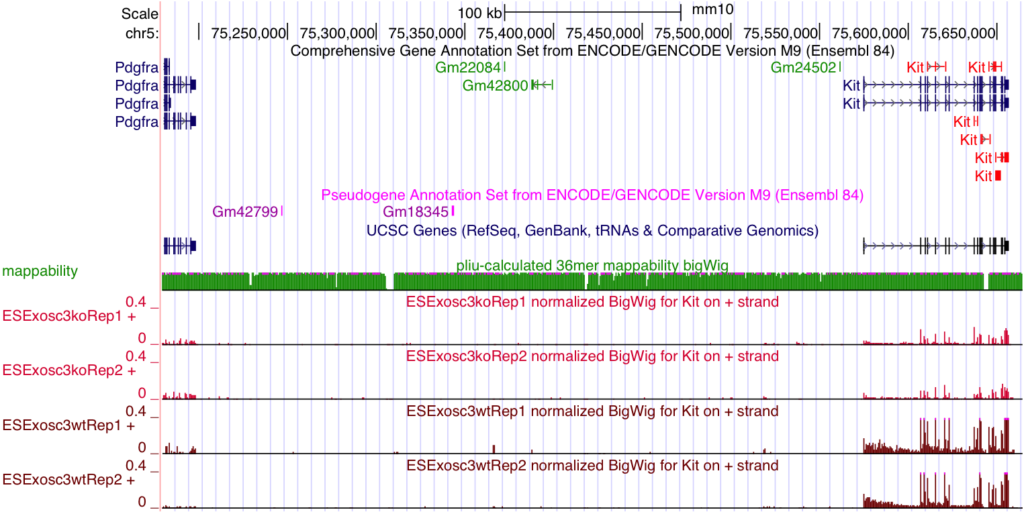
McIver SC, Katsumura KR, Davids E, Liu P, Kang YA, Yang D, Bresnick EH. Exosome complex orchestrates developmental signaling to balance proliferation and differentiation during erythropoiesis. Elife. 2016;5. doi: 10.7554/eLife.17877. PubMed PMID: 27543448; PMCID: PMC5040589.
Variant Analysis: cancer targeted sequencing panel
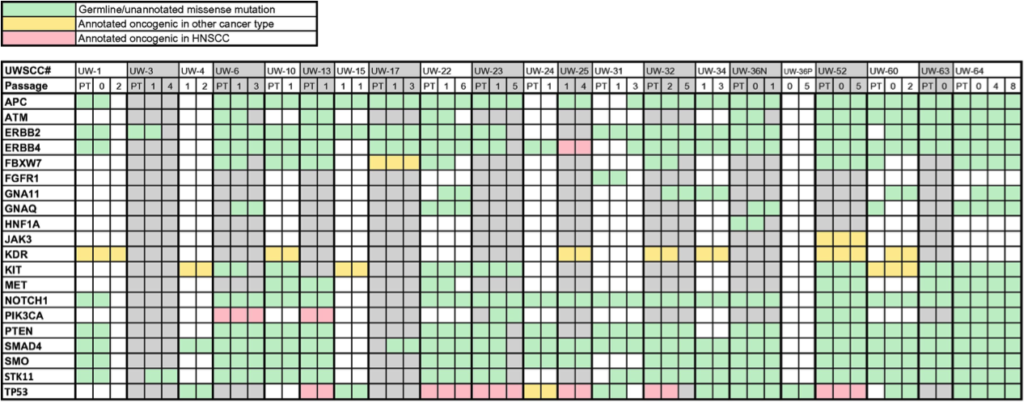

- CISR supported Randall Kimple (IR) and colleagues in validating a xenograft model for head and neck cancer.
- CISR staff scientist, Irene Ong, analyzed variants/ mutational profile of patient and xenograft samples, showing stability of grafted tumors; expanding the utility of studying tumors from head and neck cancer patients.
- Preliminary data for SPORE grant to show lab’s capability in analysis of next generation sequencing data. Three papers have been published based on these analyses.
Swick AD, Stein AP, McCulloch TM, Hartig GK, Ong IM, Sampene E, Prabakaran PJ, Liu CZ, Kimple RJ. Defining the boundaries and expanding the utility of head and neck cancer patient derived xenografts. Oral Oncol. 2017;64:65-72. doi: 10.1016/j.oraloncology.2016.11.017. PubMed PMID: 28024726; PMCID: PMC5218527.
Analysis of The Cancer Genome Atlas AML data
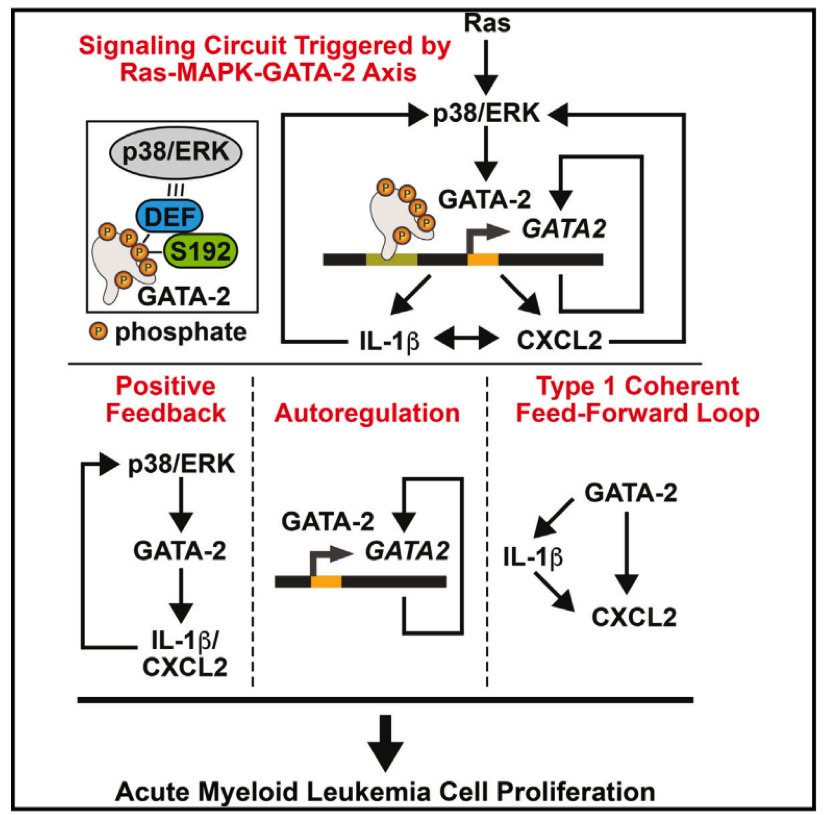
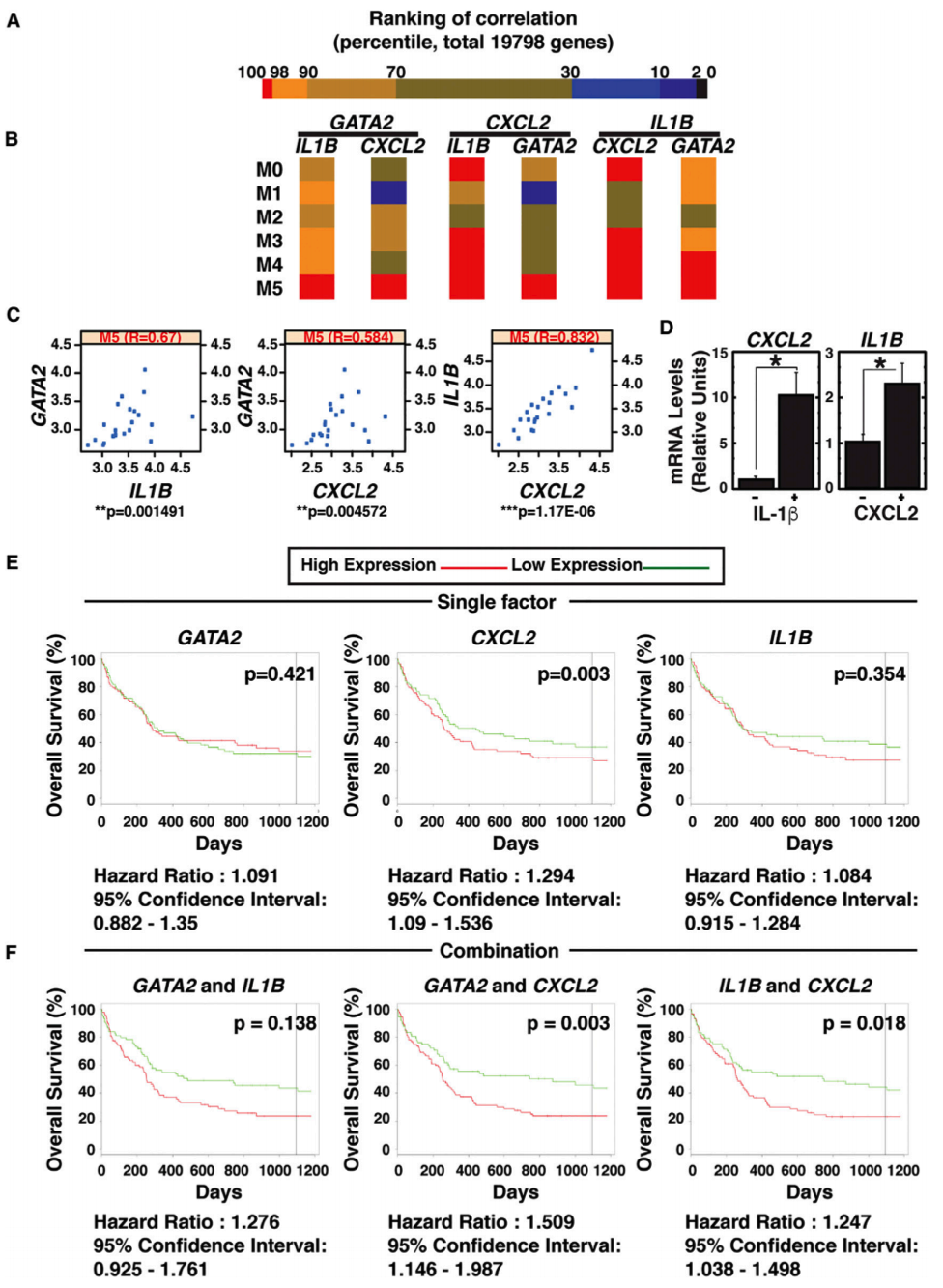
- CISR supported Emery Bresnick (GEM) and colleagues in illustrating the signaling circuit triggered by Ras-MAPK-GATA-2.
- CISR staff scientist, Irene Ong, analyzed TCGA AML data to illustrate evidence of this novel mechanism, GATA-2 regulation of IL-1β and CXCL2, in AML tumors.
- CISR also generated Kaplan-Meier curves showing GATA-2-chemokine/cytokine circuit predicts poor prognosis of an AML subtype.
Katsumura KR, OngIM, DeVilbissAW, SanalkumarR, BresnickEH. GATA Factor-Dependent Positive-Feedback Circuit in Acute Myeloid Leukemia Cells. Cell Rep. 2016;16(9):2428-41. doi: 10.1016/j.celrep.2016.07.058. PubMed PMID: 27545880; PMCID: PMC5154384.
Position Specific Scoring Matrix Motif of B'α subunit of PP2A's binding sites
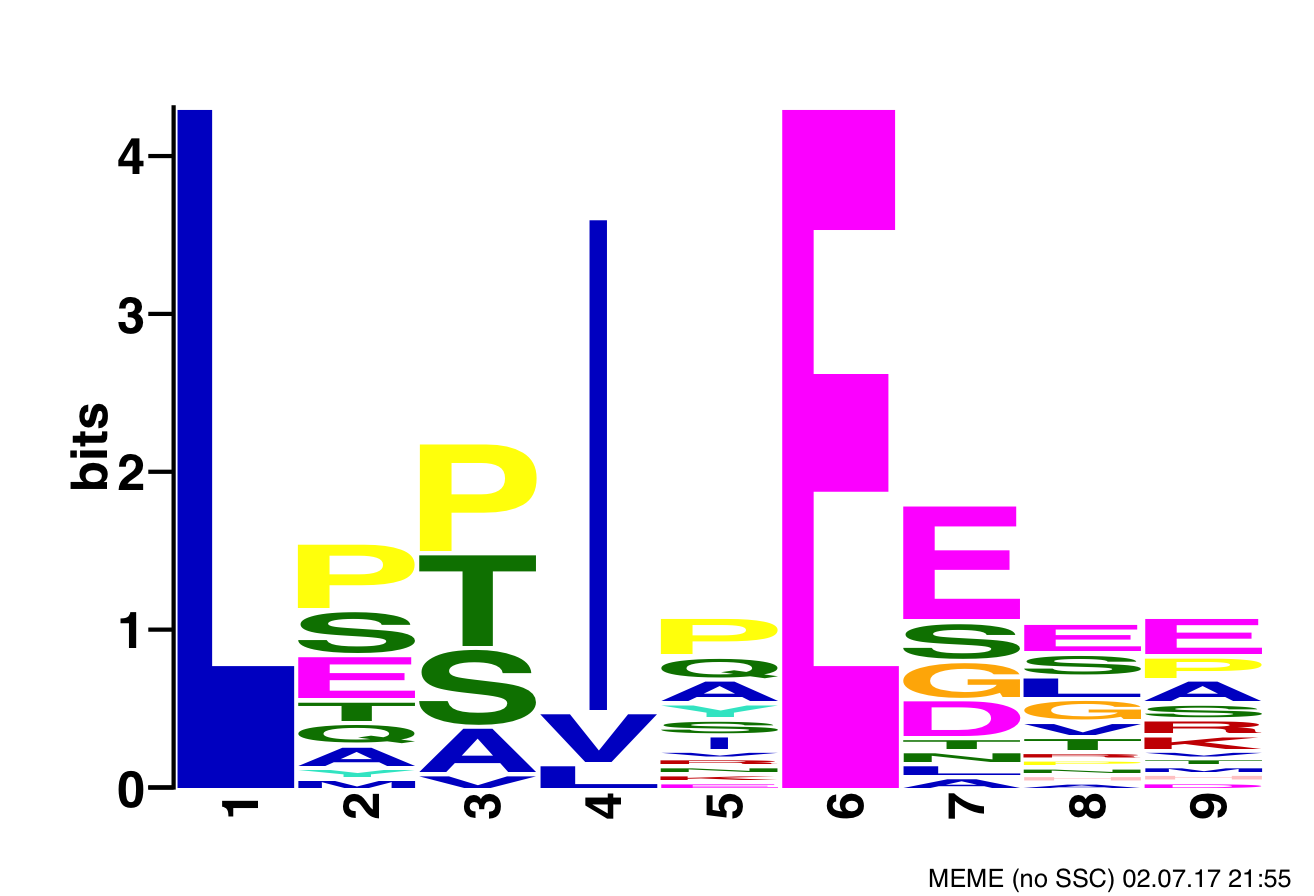
- CISR supported Dr. Xing (DT) and colleagues narrow the search space of proteins bound by B’α subunit of PP2A, which helps determine substrate recognition, regulation, and role in cytokinesis of PP2A-B’ holoenzyme, and also reduces the number of needed follow-up experiments.
- CISR staff scientist, Sean McIlwain, found consensus PP2A binding motifs within the human proteome and annotated them with secondary structures and domains, average disorder, predicted and known phosphorylation sites, and locations of the protein’s cellular super-complexes.
- CISR also performed Gene ontology analysis of the identified proteins to find enriched KEGG pathways.
Wu CG, Chen H, Guo F, Yadav VK, McIlwain SJ, Rowse M, Choudhary A, Lin Z, Li Y, Gu T, Zhen A, Xu Q, Lee W, Resch E, Johnson B, Ge Y, Ong IM, Burkard M, Ivarsson Y, Xing Y. PP2A-B’ holoenzyme substrate recognition, regulation, and role in cytokinesis. Under revision at Nature Cell Discovery.